In this section, we will build a SIR model, which is a more realistic representation of disease spread. In this model, agents can be in one of three states: Susceptible (S), Infected (I), or Recovered (R). Susceptible agents can become infected upon contact with infected agents, and infected agents can recover after a certain period, gaining immunity.
We will keep the same basic structure as before, but we will add a few more features to account for the different states of the agents. We will also implement a way to track the number of agents in each state over time, as well as visualize the results using plots.
Model Setup¶
We will start by defining the model parameters. We will need to define the infection probability, recovery probability, and the initial number of infected agents. We can use sliders to allow the user to adjust these parameters. Some of the global variables necessary for this model may be created in the Interface tab, but we will also need to define some additional global variables in the Code tab. The final interface for this simulation can be seen below:
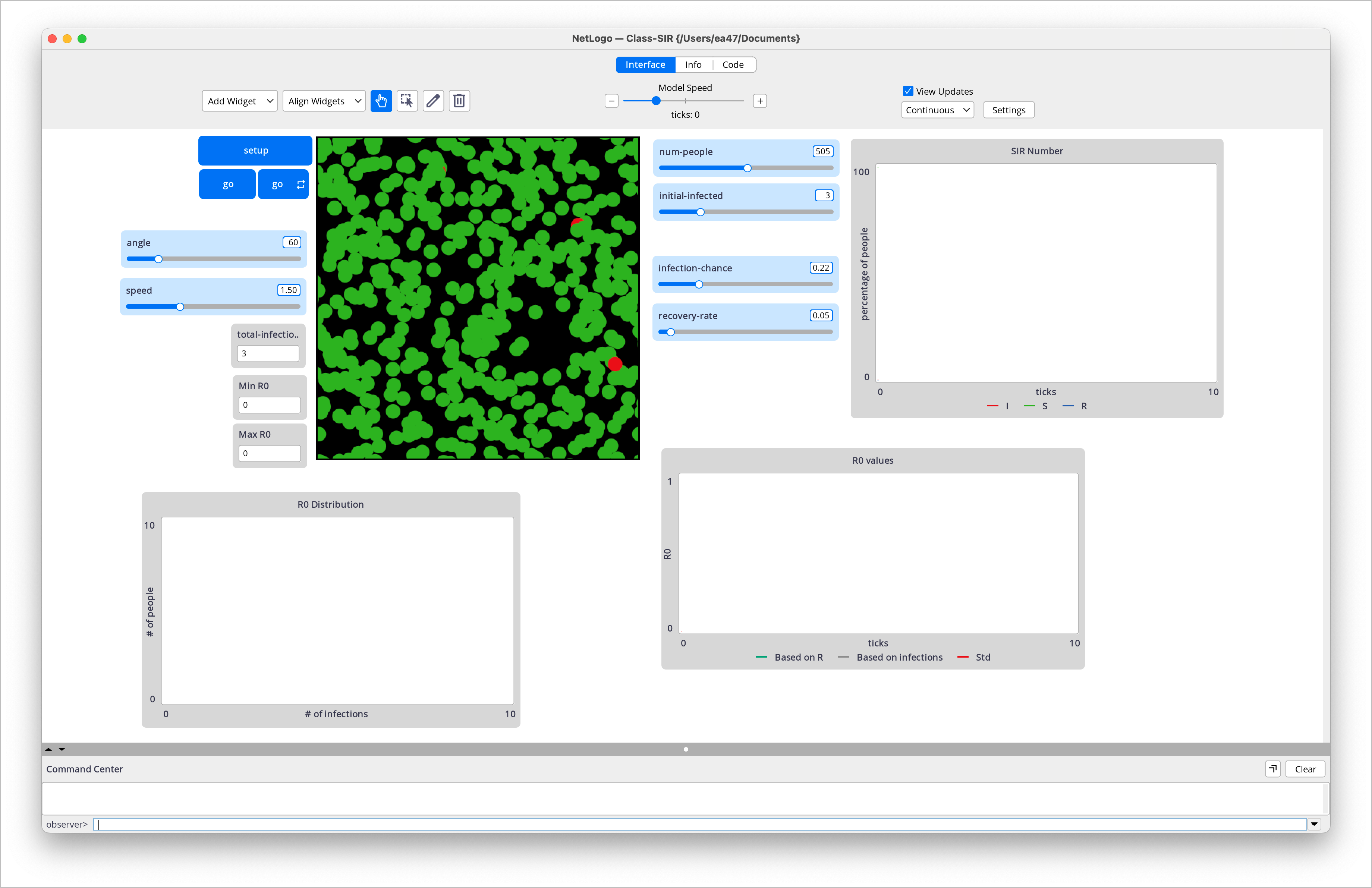
Interface for the SIR model.
globals[
infected-counts ; lists
total-infections ; lists
]
breed [people person]
people-own [
infected?
immune?
infected-by
num-infected ;; number of infections I have caused
infection-time
recovery-time
]Besides the global variables, we also added a few properties to the agents. Each agent has an infected? property to indicate if they are currently infected, an immune? property to indicate if they have recovered and are immune, an infected-by property to track who infected them, a num-infected property to count how many agents they have infected, an infection-time property to track when they got infected, and a recovery-time property to determine when they recovered.
Below we have the setup procedure, which initializes the model.
;; SETUP PROCEDURES
to setup
clear-all
create-people num-people [
set shape "circle"
set size 1.5
set color green
set infected? false
set immune? false
set infected-by nobody
set num-infected 0
set infection-time -1
set recovery-time -1
setxy random-xcor random-ycor
]
setup-infected
set infected-counts []
set total-infections initial-infected
reset-ticks
end
to setup-infected
ask n-of initial-infected people [
set infected? true
set color red
set infection-time 0
set infected-by nobody
]
endThe setup procedure initializes the model by clearing the world, creating the agents, and setting their initial properties. It also calls the setup-infected procedure to infect a certain number of agents at the start of the simulation. The infected-counts and total-infections lists are initialized to keep track of the number of infected agents over time.
Model Dynamics¶
Below is our code for the model dynamics, which includes the movement of agents, the infection process, and the recovery process.
to go
move
infect
recover-infected
if all? people [not infected?] [stop]
tick
end
to move
ask people [
left random angle
right random angle
forward speed
]
end
to infect
;; SIR model
ask people with [infected?] [
ask people-here with [not infected? and not immune?][
if random-float 1 < infection-chance [
set infected? true
set color red
set infected-by myself
set infection-time ticks
set total-infections total-infections + 1
ask myself [
set num-infected num-infected + 1
]
]
]
]
end
to recover-infected
ask people with [infected?] [
if random-float 1 < recovery-rate [
set infected? false
set recovery-time ticks
set immune? true
set color blue
]
]
endThe go procedure is called at each time step and calls the move, infect, and recover-infected procedures. It also checks if there are any infected agents left; if not, it stops the simulation.
The move procedure updates the position of each agent based on its heading and speed. The infect procedure implements the infection process, where infected agents can infect susceptible agents in the same location with a certain probability. The recover-infected procedure implements the recovery process, where infected agents can recover and become immune with a certain probability.
Data Collection and Visualization¶
We are interested in knowing how the number of infected agents changes over time, as well as the values for R0 (R-naught) and the distribution of infections caused by each agent. We can use the infected-counts and total-infections lists to keep track of the number of infected agents over time and the total number of infections, respectively. We can also calculate R0 as the average number of infections caused by each infected agent.
Let’s start by calculating R0 based on the number of recovered agents and the number of infections they caused. We can create a reporter called calculate-R0 to do this.
to-report calculate-R0
;; from the immune people (those who have recovered)
let recovered-people people with [immune?]
ifelse any? recovered-people [
;; there are people in the R compartment
report mean [num-infected] of recovered-people
][
;; agentset is empty
report 0
]
endNext, we can create a reporter called current-mean-R0 to calculate the current average R0 across all individuals who have infected at least one person.
to-report current-mean-R0
;; current average R0 across all individuals who have infected at least one person
let infectors people with [num-infected > 0]
ifelse any? infectors[
report mean [num-infected] of infectors
][
;; no one infected anyone yet
report 0
]
endThe next two reports, max-R0 and min-R0-nonzero, will help us track the maximum and minimum non-zero R0 values among individuals who have infected others. That is, the maximum and minimum number of people infected by a single individual.
to-report max-R0
;; maximum number of people infected by a single individual
let infectors people with [num-infected > 0]
ifelse any? infectors [
report max [num-infected] of infectors
][
report 0
]
end
to-report min-R0-nonzero
; minimum non-zero R0 value
let infectors people with [num-infected > 0]
ifelse any? infectors [
report min [num-infected] of infectors
][
report 0
]
endWe can also create a reporter called R0-distribution to return a list of all num-infected values for analysis. That will be used to create a histogram of the distribution of infections caused by each agent.
to-report R0-distribution
; return a list of all num-infected values for analysis
report [num-infected] of people with [num-infected > 0]
endOther stats are also interesting to track, such as the standard deviation of R0 values. We can create a reporter called R0-standard-deviation to calculate that.
to-report R0-standard-deviation
; standard deviation of R0 values
let infectors people with [num-infected > 0]
ifelse any? infectors [
report standard-deviation [num-infected] of infectors
][
report 0
]
endFinally, we can create reporters to calculate the percentage of agents in each state (Susceptible, Infected, Recovered) at any given time. These reporters will be useful for plotting the SIR curves.
to-report percent-infected
report (count people with [infected?] / num-people) * 100
end
to-report percent-recovered
report (count people with [immune?] / num-people) * 100
end
to-report percent-susceptible
report (count people with [not infected? and not immune?] / num-people) * 100
endAll these reporters can be used to create plots and visualizations in the Interface tab. For example, we can create a plot to show the SIR curves over time, with the x-axis representing time (ticks) and the y-axis representing the percentage of agents in each state. We can also create a histogram to show the distribution of infections caused by each agent.
You can investigate the plots and monitors by downloading and playing with our model here.